A new study from researchers at the Harvard School of Public Health shows that the microbial communities each of us carries on our skin or in our bodies can be used to uniquely identify individuals, much like a fingerprint. The team behind the study showed that personal microbiomes contain enough distinguishing features to identify a person over a long amount of time from a research study population of hundreds of people. It is the first that shows microbiome identification is feasible.
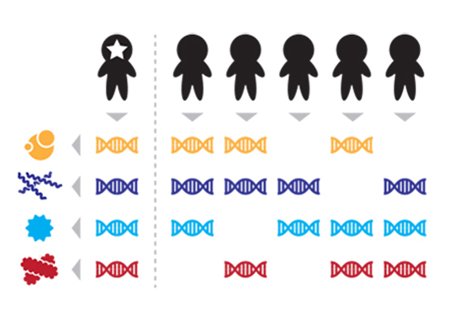 Taken together, DNA sequences from four microbial species distinguish the starred person’s microbiome from those of five other people
Taken together, DNA sequences from four microbial species distinguish the starred person’s microbiome from those of five other people
Image via hsph.harvard.edu
“Linking a DNA sample to a database of DNA ‘fingerprints’ is the basis for forensic genetics, now a decades-old field.”
“We’ve shown that the same sort of linking is possible using DNA sequences from microbes inhabiting the human body — no human DNA required. This opens the door to connecting human microbiome samples between databases, which has the potential to expose sensitive subject information — for example, a sexually-transmitted infection, detectable from the microbiome sample itself,” said lead author Eric Franzosa, research fellow in the Department of Biostatistics at Harvard Chan.
More than just a gut feeling – it’s gut forensics
The team used publicly available microbiome data mined from the Human Microbiome Project (HMP), which surveyed microbe samples from stool, saliva, skin and other sites from up to 242 individuals over a month-long period. They adapted a computer algorithm to crunch the data and obtained combinations of stable and distinguishing sequence features from the initial samples that work as individual-specific “codes”.
When compared to microbiome samples collected at follow-up visits with the same individuals and to samples from independent groups of individuals, the codes were shown to be unique among hundreds of individuals. A large fraction of the microbial signature remained stable over a one-year sampling period, those obtained from gut samples in particular. Here, more than 80% of individuals could be identified without a trace of doubt for up to a year after the sampling period.
“Although the potential for any data privacy concerns from purely microbial DNA is very low, it’s important for researchers to know that such issues are theoretically possible,” said senior author Curtis Huttenhower, associate professor of computational biology and bioinformatics at Harvard Chan School. “Perhaps even more exciting are the implications of the study for microbial ecology, since it suggests our unique microbial residents are tuned to the environment of our body — our genetics, diet, and developmental history — in such a way that they stick with us and help to fend off less-friendly microbial invaders over time.”



